5. Data formats¶
This section gives a description of the raw data produced by PANIC and how they are organized. However, for a deeper description, see the GEIRS manual.
5.1. Detector¶
The PANIC instrument had initially was mounted with a mosaic of four 2k x 2k HAWAII-2RG detectors. However, during 2020 a new single (monolithic) 4k x 4k HAWAII-4RG detector was installed due to the damages in the HAWAII-2RG.
H2RG: The detector is an array (FPA) of four HAWAII-2RG detectors. The inter-chip gap between the detectors is only 167 pixels (or 75 arcsec at the 2.2m telescope) and is filled by dithering with sufficient amplitude. For applications which image only 30x30 arcmin this design is ideal.
HAWAII-2RG detectors have an effective surface of 2040x2040 sensitive pixels. A 4-pixel wide border is used as reference to correct for relatively slow bias drifts.
H4RG: The HAWAII-4RG detector is a 4096x4096 pixels with 15um pixel pitch; it is the next generation, state-of-the-art readout integrated circuit for visible and infrared instrumentation in ground-based and space-based telescope applications.
5.2. FITS¶
GEIRS, the software part in charge of the data acquisition and saving, is capable of saving the frames in different FITS (Flexible Image Transport System) formats (integrated, FITS-cubes, MEF, etc ). Next ones are available in the Observation Tool (OT) when an OP (Observing Program) is defined:
Multi-Extension FITS (MEF) - Integrated
Multi-Extension FITS (MEF) - Cube
Integrated All (SEF - Integrated)
FITS Cube (SEF - Cube)
Individual (SEF - Individual)
However, PAPI does not accept any kind of FITS data files available in GEIRS, only the configured in the OT, except Individual. As result, PAPI accepts the next type of FITS files (in order of preference):
Integrated Multi-Extension-FITS (MEF): a unique FITS file with four extensions (MEF), where each extension corresponds to one of the 4 images produced by the single detector chips. If the number of coadd (NCOADDS) is > 0, then they will be integrated (arithmetic sum) in a single image. This is the default and more common saving mode used; in fact, it is the default and more wished saving mode. This mode will also be used when the software or hardware sub-windowing is set and the integrated option is selected. Then, there will be an extension for each sub-window. (This mode makes sense only for the 4xH2RG)
Non-integrated Multi-Extension-FITS (MEF): a unique FITS file with four extensions (MEF), one per each detector (or window), having each extension N planes, where N is the number of coadds (NCOADDS), ie. a cube of N planes. This mode will be also used when the software or hardware subwindowing is set up and the no-integrated option is selected. (This mode makes sense only for the 4xH2RG)
Single integrated FITS file: the four H2RG detectors / single monolithic H4RG are/is saved in single file and in a single extension FITS image (SEF). If the number of coadds (NCOADDS) is > 0, then they are integrated (arithmetic sum) in a single frame.
Single non-integrated FITS-cube: the four detectors/single monolithic H4RG are/is saved in a single extension FITS (SEF) file, and each individual exposition in a plane/layer of a cube. It means N planes, where N is the number of coadds or expositions.
Note
Currently PAPI is not working with non-integrated individual files of an exposition. In case you are interested in no-integrated files and wish to reduce the data with PAPI, you should use SEF or MEF non-integrated FITS-cube mode.
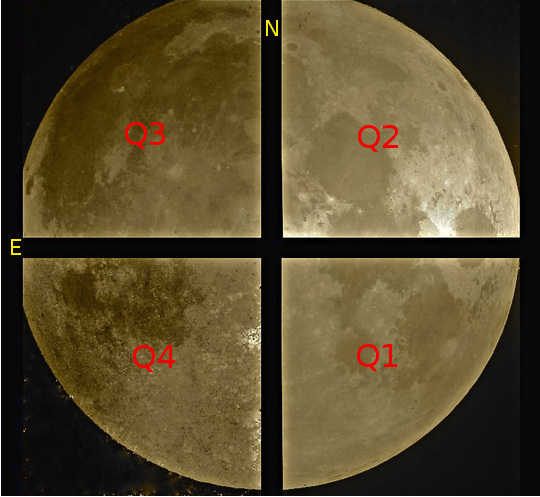
Beware that the order of the chip in the raw image produced is as described in next figure:
5.2.1. HAWAII-4RG¶
Hawaii-4RG is a monolithic IR detector, therefore neither quadrants nor MEF files do not make sense in that case.
5.2.1.1. H4RG Header¶
The header keywords currently used in a raw H4RG PANIC FITS file is as shown bellow:
SIMPLE = T
BITPIX = 32
NAXIS = 2 / 2
NAXIS1 = 4096
NAXIS2 = 4096
EXTEND = T / FITS dataset may contain extensions
COMMENT FITS (Flexible Image Transport System) format is defined in 'Astronomy
COMMENT and Astrophysics', volume 376, page 359; bibcode: 2001A&A...376..359H
BSCALE = 1.
BZERO = 0. / [adu] real = bzero + bscale*value
BUNIT = 'adu ' / [adu]
MJD-OBS = 59698.561326 / [d] Modified Julian Date of DATE-OBS
DATE-OBS= '2022-04-29T13:27:09.3989' / [d] UTC end of first frame read
DATE = '2022-04-29T13:33:13' / file creation date (YYYY-MM-DDThh:mm:ss UT)
UT = 48429.398901 / [s] 13:27:09.3989 UTC at end of first frame rea
LST = 16354.189459 / [s] local sidereal time: 04:32:34 (EOread)
ALT = 55.413811 / [deg] Altitude of the object above horizon
AZ = 111.106918 / [deg] Azimuth (S=0, W=+90) of the object
PARANG = 68.593735 / [deg] Parallactic angle
ORIGIN = 'Centro Astronomico Hispano Aleman (CAHA)'
OBSERVER= 'my_observer'
TELESCOP= 'CA-3.5 '
FRATIO = 'F/10 ' / [1]
OBSGEO-L= 8.724 / [deg] telesc. geodet. longit. 2015A&A..574A..36
OBSGEO-B= 49.396 / [deg] telesc. geodet. latit. 2015A&A..574A..36R
OBSGEO-H= 560. / [m] above sea level 2015A&A..574A..36R
LAMPSTS = ' ' / calib. lamp
INSTRUME= 'PANIC ' / PAnoramic Near Infrared camera for Calar Alto
CAMERA = 'HgCdTe IR-Camera (1 H4RGs)'
PIXSCALE= 0.186667 / [arcsec/pix]
EGAIN1 = 4. / [e-/adu] electrons/DN
ENOISE1 = 16. / [e-] electrons/read ems=1
ROVER = 'MPIA IR-ROelectronic Vers. 3.1' / Version det. electronics
WPOS = 5 / [ct] number of GEIRS wheels
W1POS = 'Coldstop22'
W2POS = 'Blank '
W3POS = 'Blank '
W4POS = 'Blank '
W5POS = 'Blank '
FILTER = 'NO ' / filter macro name of filter combinations
STRT_INT= 48429.398901 / [s] 13:27:09.3989 start integration (UT)
STOP_INT= 48463.913569 / [s] 13:27:43.9136 stop integration (UT)
RA = 13.856732 / [deg] R.A.: 00:55:25.6
DEC = 49.296 / [deg] Dec.: +49:17:46
EQUINOX = 2000. / [a] Julian Epoch
AIRMASS = 1.214279 / [1] airmass
HA = 54.29408 / [deg] H.A. '03:37:10.58'
T_FOCUS = 0. / [mm] telescope focus
CASSPOS = 0. / [deg] cassegrain position rel. to NSEW
OBJECT = 'J005525+491745' / telescope target
POINT_NO= 0 / [ct] pointing counter
DITH_NO = 0 / [ct] dither step
EXPO_NO = 10 / [ct] exposure/read counter
FILENAME= 'OPT_FOCUS_DARK_01_0001.fits'
FILE_ID = 'Panic.2022-04-29T13:27:09.398_0001_001' / instru., time, image, windo
TPLNAME = ' ' / macro/template name
NQCHAN = 64 / [ct] output channels of each Hawaii chip
PTIME = 2 / pixel-time-base index
PREAD = 10000 / [ns] pixel read selection
PSKIP = 150 / [ns] pixel skip selection
LSKIP = 201 / [ns] line skip selection
READMODE= 'continuous.sampling.read' / read cycle-type
IDLEMODE= 'wait ' / idle to read transition
IDLETYPE= 'ReadWoConv' / idle cycle-type
SAVEMODE= 'continuous.sampling.read' / save cycle-type
NEXP = 6 / cycle repeat count
CPAR1 = 2 / cycle type parameter
ITIME = 2.765469 / [s] scheduled integration time
CTIME = 5.530939 / [s] read-mode cycle time
EMSAMP = 1 / [ct] electronic multi-sampling
NCOADDS = 6 / [ct] coadds (total)
EXPTIME = 16.592816 / [s] total integ. time
FRAMENUM= 1 / sum of 6 image(s)
SKYFRAME= 'unknown '
DETSEC = '[1:4096,1:4096]' / [pix] xrange and yrange of window
DATASEC = '[1:4096,1:4096]' / [pix] xy-range of science data
DETSIZE = '[1:4096,1:4096]' / [px] full size of the 4 detector mosaic
CHIPSIZX= 4096 / [pix] single chip pixels in x
CHIPSIZY= 4096 / [pix] single chip pixels in y
HINVDIR = 0 / left-right directions of horiz. scanner
VINVDIR = 0 / top-bottom directions of vert. scanner
DETROT90= 2 / [ct] 90 deg SW image cw rotations
DETXYFLI= 0 / [1] SW image flip (1=RightLeft, 2=UpDown)
B_EXT1 = 2.080078 / [V] external bias 2130
B_DSUB1 = 0.600179 / [V] det. bias voltage DSUB 1034
B_VREST1= 0.30009 / [V] det. bias voltage VRESET 517
B_VBIAG1= 2.199707 / [V] det. bias voltage VBIASGATE 3604
B_VNBIA1= 0. / [V] det. bias voltage VNBIAS 0
B_VPBIA1= 0. / [V] det. bias voltage VPBIAS 0
B_VNCAS1= 0. / [V] det. bias voltage VNCASC 0
B_VPCAS1= 0. / [V] det. bias voltage VPCASC 0
B_VBOUB1= 0. / [V] det. bias voltage VBIASOUTBUF 0
B_REFSA1= 0. / [V] det. bias voltage REFSAMPLE 0
B_REFCB1= 0. / [V] det. bias voltage REFCOLBUF 0
TEMP_A = 79.331001 / [K] Moly frame (-193.82 C)
TEMP_B = 79.329002 / [K] Detector (-194 C)
PRESS1 = 4.0E-05 / [Pa] (4.000e-10 bar) , 'pressure1'
TEMPMON = 8 / [ct] # of temp. monitrd 2022-04-29 13:30 loc. t
TEMPMON1= 99.503998 / [K] (-173.65 C) Cold plate
TEMPMON2= 108.75 / [K] (-164.40 C) Lens Mount 1
TEMPMON3= 102.010002 / [K] (-171.14 C) Charcoal
TEMPMON4= 77.526001 / [K] (-195.62 C) LN2 detector tank
TEMPMON5= 104.050003 / [K] (-169.10 C) Filter wheel housing
TEMPMON6= 105.139999 / [K] (-168.01 C) Preamps
TEMPMON7= 81.721001 / [K] (-191.43 C) LN2 main tank
TEMPMON8= 103.540001 / [K] (-169.61 C) Radiation shield
CREATOR = 'GEIRS : trunk-r799M-65 (Apr 7 2022, 14:12:56)'
PLX_API = 8.23 / Major and Minor PLX API version
OS = 'Linux irws1 5.3.18-150300.59.63-default #1 SMP Tue Apr 5 12:47:31 UT'
UUID = 'e931efc4-c7c0-11ec-a4f3-90b11c480ad2' / Universally unique identifier
COMMENT = 'no comment'
OBSERVAT= 'CAHA ' / Calar Alto, Andalucia, Spain, panic.caha.es
CHIPID = '19908 ' / detect. HW ID
OPCYCL = 10 / Operation cycle number
OPDATE = '2016-02-18T15:55:00' / UT-date of operation cycle start
MNTCYCL = 20 / Mounting cycle number
MNTDATE = '2017-01-18T15:47:00' / UT-date of mounting cycle start
CUNIT1 = 'deg ' / WCS units along axis 1
CUNIT2 = 'deg ' / WCS units along axis 2
CTYPE1 = 'RA---TAN' / WCS axis 1
CTYPE2 = 'DEC--TAN' / WCS axis 2
CRVAL1 = 13.8567324535148 / [deg] RA in center
CRVAL2 = 49.296 / [deg] DEC in center
CD1_1 = -5.18518520726098E-05 / [deg/px] WCS matrix diagonal
CD2_2 = 5.18518520726098E-05 / [deg/px] WCS matrix diagonal
CD1_2 = -1.0634992634962E-14 / [deg/px] WCS matrix outer diagonal
CD2_1 = -1.0634992634962E-14 / [deg/px] WCS matrix outer diagonal
CRPIX1 = 2049 / [px] RA and DEC center along axis 1
CRPIX2 = 2049 / [px] RA and DEC center along axis 2
EOF00000= 48429.398866 / [s] 13:27:09.3988 UTC past midnight
EOF00001= 48433.443542 / [s] 13:27:13.4435 +4.04468 UTC past midnight
EOF00002= 48436.208914 / [s] 13:27:16.2089 +2.76537 UTC past midnight
EOF00003= 48438.974351 / [s] 13:27:18.9743 +2.76544 UTC past midnight
EOF00004= 48441.739649 / [s] 13:27:21.7396 +2.7653 UTC past midnight
EOF00005= 48444.506323 / [s] 13:27:24.5063 +2.76667 UTC past midnight
EOF00006= 48447.270818 / [s] 13:27:27.2708 +2.76449 UTC past midnight
EOF00007= 48450.036261 / [s] 13:27:30.0362 +2.76544 UTC past midnight
EOF00008= 48452.801598 / [s] 13:27:32.8015 +2.76534 UTC past midnight
EOF00009= 48455.567247 / [s] 13:27:35.5672 +2.76565 UTC past midnight
EOF00010= 48458.332675 / [s] 13:27:38.3326 +2.76543 UTC past midnight
EOF00011= 48461.098155 / [s] 13:27:41.0981 +2.76548 UTC past midnight
EOF00012= 48463.863639 / [s] 13:27:43.8636 +2.76548 UTC past midnight
OBS_TOOL= 'createDS' / PANIC Observing Tool Software version
PROG_ID = ' ' / PANIC Observing Program ID
OB_ID = '1 ' / PANIC Observing Block ID
OB_NAME = 'OB_dummy' / PANIC Observing Block Name
OB_PAT = 'unknown ' / PANIC Observing Block Pattern Type
PAT_NAME= 'unknown ' / PANIC Observing Sequence Pattern Name
PAT_EXPN= 1 / PANIC Observing Exposition Number
PAT_NEXP= 7 / PANIC Observing Number of Expositions
IMAGETYP= 'DARK ' / PANIC Image type
END
5.2.2. HAWAII-2RG¶
Next table shows the mapping of extension/quadrant names and detectors with the H2RG detector array:
Extension Name |
Q1 |
Q2 |
Q3 |
Q4 |
|---|---|---|---|---|
Detector Hw ID |
SG1 |
SG2 |
SG3 |
SG4 |
Note that the order of the extensions in the FITS file is Q1 (ext. 1), Q2 (ext. 2), Q3 (ext. 3) and Q4 (ext. 4).
Note
The above extension name and order are only valid from version GEIRS-r731M-18 onwards.
5.2.2.1. H2RG Header¶
The header keywords currently used in a raw H2RG PANIC FITS file is as shown bellow:
Primary Header
SIMPLE = T
BITPIX = 32
NAXIS = 2 / 2
NAXIS1 = 4096
NAXIS2 = 4096
EXTEND = T / FITS dataset may contain extensions
COMMENT FITS (Flexible Image Transport System) format is defined in 'Astronomy
COMMENT and Astrophysics', volume 376, page 359; bibcode: 2001A&A...376..359H
BSCALE = 1.
BZERO = 0. / [adu] real = bzero + bscale*value
BUNIT = 'adu ' / [adu]
MJD-OBS = 57170.68257 / [d] Modified julian date 'days' of observation
DATE-OBS= '2015-05-28T16:22:54.0402' / [d] UT-date of observation end
DATE = '2015-05-28T16:22:54' / file creation date (YYYY-MM-DDThh:mm:ss UT)
UT = 58974.040247 / [s] 16:22:54.0402 UTC at EOread
LST = 30949.087329 / [s] local sidereal time: 08:35:49.087 (EOread)
ORIGIN = 'Centro Astronomico Hispano Aleman (CAHA)'
OBSERVER= 'Mathar '
TELESCOP= 'CA-2.2 '
FRATIO = 'F/08 ' / [1]
OBSGEO-L= -2.546135 / [deg] telescope geograph. longit. 2015A&A..574A
OBSGEO-B= 37.223037 / [deg] telescope geograph. latit. 2015A&A..574A.
OBSGEO-H= 2168. / [m] above sea level 2015A&A..574A..36R
LAMPSTS = ' ' / calib. lamp
INSTRUME= 'PANIC ' / PAnoramic Near Infrared camera for Calar Alto
CAMERA = 'HgCdTe (4096x4096) IR-Camera (4 H2RGs)'
PIXSCALE= 0.45 / [arcsec/px]
EGAIN1 = 4.84 / [ct] electrons/DN
EGAIN2 = 4.99 / [ct] electrons/DN
EGAIN3 = 5.02 / [ct] electrons/DN
EGAIN4 = 5.45 / [ct] electrons/DN
ENOISE1 = 16. / [ct] electrons/read ems=1
ENOISE2 = 16.6 / [ct] electrons/read ems=1
ENOISE3 = 18.5 / [ct] electrons/read ems=1
ENOISE4 = 17.9 / [ct] electrons/read ems=1
ROVER = 'MPIA IR-ROelectronic Vers. 3' / Version det. electronics
WPOS = 5 / [ct] number of GEIRS wheels
W1POS = 'Coldstop22'
W2POS = 'H '
W3POS = 'Ks '
W4POS = 'dummy '
W5POS = 'Black '
FILTER = 'NO ' / filter macro name of filter combinations
STRT_INT= 58943.164225 / [s] 16:22:23.1642 start integration (UT)
STOP_INT= 58946.502476 / [s] 16:22:26.5025 stop integration (UT)
RA = 172.8182 / [deg] R.A.: 11:31:16.4
DEC = 33.088802 / [deg] Dec.: +33:05:20
EQUINOX = 2000. / [a] Julian Epoch
OBSEPOCH= 2015.403645 / [a] Julian Epoch
AIRMASS = 1.232127 / [1] airmass
HA = 316.144687 / [deg] H.A. '21:04:34.72'
T_FOCUS = 0. / [mm] telescope focus
CASSPOS = 0. / [deg] cassegrain position rel. to NSEW
OBJECT = 'unknown ' / telescope target
POINT_NO= 0 / [ct] pointing counter
DITH_NO = 0 / [ct] dither step
EXPO_NO = 2 / [ct] exposure/read counter
FILENAME= 'test_0001.fits'
FILE_ID = 'Panic.2015-05-28T16:22:23.164_0001_001' / instru., time, image, windo
TPLNAME = ' ' / macro/template name
TIMER0 = 2740 / [ms]
TIMER1 = 2740 / [ms]
TIMER2 = 0 / [us]
PTIME = 2 / pixel-time-base index
PREAD = 10000 / [ns] pixel read selection
PSKIP = 150 / [ns] pixel skip selection
LSKIP = 150 / [ns] line skip selection
READMODE= 'line.interlaced.read' / read cycle-type
IDLEMODE= 'wait ' / idle to read transition
IDLETYPE= 'ReadWoConv' / idle cycle-type
SAVEMODE= 'line.interlaced.read' / save cycle-type
NEXP = 1 / cycle repeat count
CPAR1 = 1 / cycle type parameter
ITIME = 2.739931 / [s] (on chip) integration time
CTIME = 5.481201 / [s] read-mode cycle time
HCOADDS = 1 / [ct] # of hardware coadds
EMSAMP = 1 / [ct] electronic multi-sampling
PCOADDS = 1 / [ct] # of coadded plateaus/periods
SCOADDS = 1 / [ct] # of software coadds
SWMSAMP = 1 / [ct] # software multisampling
NCOADDS = 1 / [ct] effective coadds (total)
EXPTIME = 2.739931 / [s] total integ. time
FRAMENUM= 1 / of 1 saved
SKYFRAME= 'unknown '
DETSEC = '[1:4096,1:4096]' / [px] xrange and yrange of window
DATASEC = ' ' / [px] xy-range of science data
DETSIZE = '[1:4096,1:4096]' / [px] full size of the 4 detector mosaic
CHIPSIZX= 2048 / [px] single chip pixels in x
CHIPSIZY= 2048 / [px] single chip pixels in y
DETROT90= 0 / [ct] 90 deg SW image cw rotations
DETXYFLI= 0 / [1] SW image flip (1=RightLeft, 2=UpDown)
B_EXT1 = 2.679688 / [V] external bias 2744
B_EXT2 = 2.679688 / [V] external bias 2744
B_EXT3 = 2.679688 / [V] external bias 2744
B_EXT4 = 2.679688 / [V] external bias 2744
B_DSUB1 = 1.569727 / [V] det. bias voltage DSUB 3420
B_DSUB2 = 1.569727 / [V] det. bias voltage DSUB 3420
B_DSUB3 = 1.569727 / [V] det. bias voltage DSUB 3420
B_DSUB4 = 1.569727 / [V] det. bias voltage DSUB 3420
B_VREST1= 1.07999 / [V] det. bias voltage VRESET 2353
B_VREST2= 1.07999 / [V] det. bias voltage VRESET 2353
B_VREST3= 1.07999 / [V] det. bias voltage VRESET 2353
B_VREST4= 1.07999 / [V] det. bias voltage VRESET 2353
B_VBIAG1= 2.199707 / [V] det. bias voltage VBIASGATE 3604
B_VBIAG2= 2.199707 / [V] det. bias voltage VBIASGATE 3604
B_VBIAG3= 2.199707 / [V] det. bias voltage VBIASGATE 3604
B_VBIAG4= 2.199707 / [V] det. bias voltage VBIASGATE 3604
B_VNBIA1= 0. / [V] det. bias voltage VNBIAS 0
B_VNBIA2= 0. / [V] det. bias voltage VNBIAS 0
B_VNBIA3= 0. / [V] det. bias voltage VNBIAS 0
B_VNBIA4= 0. / [V] det. bias voltage VNBIAS 0
B_VPBIA1= 0. / [V] det. bias voltage VPBIAS 0
B_VPBIA2= 0. / [V] det. bias voltage VPBIAS 0
B_VPBIA3= 0. / [V] det. bias voltage VPBIAS 0
B_VPBIA4= 0. / [V] det. bias voltage VPBIAS 0
B_VNCAS1= 0. / [V] det. bias voltage VNCASC 0
B_VNCAS2= 0. / [V] det. bias voltage VNCASC 0
B_VNCAS3= 0. / [V] det. bias voltage VNCASC 0
B_VNCAS4= 0. / [V] det. bias voltage VNCASC 0
B_VPCAS1= 0. / [V] det. bias voltage VPCASC 0
B_VPCAS2= 0. / [V] det. bias voltage VPCASC 0
B_VPCAS3= 0. / [V] det. bias voltage VPCASC 0
B_VPCAS4= 0. / [V] det. bias voltage VPCASC 0
B_VBOUB1= 0. / [V] det. bias voltage VBIASOUTBUF 0
B_VBOUB2= 0. / [V] det. bias voltage VBIASOUTBUF 0
B_VBOUB3= 0. / [V] det. bias voltage VBIASOUTBUF 0
B_VBOUB4= 0. / [V] det. bias voltage VBIASOUTBUF 0
B_REFSA1= 0. / [V] det. bias voltage REFSAMPLE 0
B_REFSA2= 0. / [V] det. bias voltage REFSAMPLE 0
B_REFSA3= 0. / [V] det. bias voltage REFSAMPLE 0
B_REFSA4= 0. / [V] det. bias voltage REFSAMPLE 0
B_REFCB1= 0. / [V] det. bias voltage REFCOLBUF 0
B_REFCB2= 0. / [V] det. bias voltage REFCOLBUF 0
B_REFCB3= 0. / [V] det. bias voltage REFCOLBUF 0
B_REFCB4= 0. / [V] det. bias voltage REFCOLBUF 0
TEMP_A = 79.068001 / [K] Moly frame (-194.08 C)
TEMP_B = 79.999001 / [K] Detector (-193 C)
PRESS1 = 1.0E-05 / [Pa] (1.020e-10 bar) , 'pressure1'
TEMPMON = 8 / [ct] # of temp. monitrd 2015-05-28 16:21 loc. t
TEMPMON1= 84.508003 / [K] (-188.64 C) Cold plate
TEMPMON2= 97.056 / [K] (-176.09 C) Lens Mount 1
TEMPMON3= 85.961998 / [K] (-187.19 C) Charcoal
TEMPMON4= 75.846001 / [K] (-197.30 C) LN2 detector tank
TEMPMON5= 87.633003 / [K] (-185.52 C) Filter wheel housing
TEMPMON6= 94.026001 / [K] (-179.12 C) Preamps
TEMPMON7= 79.591003 / [K] (-193.56 C) LN2 main tank
TEMPMON8= 89.347 / [K] (-183.80 C) Radiation shield
CREATOR = 'GEIRS : trunk-r737M-13 (May 28 2015, 16:17:00), Panic'
COMMENT = 'no comment'
OBSERVAT= 'CAHA ' / Calar Alto, Almeria, Andalucia, Spain, panic.ca
OPCYCL = 9 / Operation cycle number
OPDATE = '2015-04-28T15:16:00' / UT-date of operation cycle start
MNTCYCL = 7 / Mounting cycle number
MNTDATE = '2015-01-29T15:00:00' / UT-date of mounting cycle start
HIERARCH CAHA AMBI WINSP = 4.5 / [m/s] wind speed day=20150528 UT=1448
HIERARCH CAHA AMBI WINDIR = 149. / [deg] wind direction day=20150528 UT=1448
HIERARCH CAHA AMBI TEMP = 15.2 / [C] temperature day=20150528 UT=1448
HIERARCH CAHA AMBI HUMI = 46 / [%] rel. humidity day=20150528 UT=1448
HIERARCH CAHA AMBI DEWP = 3.7 / [C] dew point day=20150528 UT=1448
HIERARCH CAHA AMBI PRESS = 778. / [hPa] air pressure day=20150528 UT=1448
HIERARCH CAHA AMBI CLOUD = -25.7 / [] cloud sensor day=20150528 UT=1448
COMMENT Linux panic22 3.11.6-4-desktop #1 SMP PREEMPT Wed Oct 30 18:04:56 UTC 20
COMMENT 13 (e6d4a27) x86_64
COMMENT Plx API Version 7.10
EOFRM000= 58943.164227 / [s] 16:22:23.1642 UTC past midnight
EOFRM002= 58944.177113 / [s] 16:22:24.1771 +1.01289 UTC past midnight
END
Extensions Header (SG1)
XTENSION= 'IMAGE ' / IMAGE extension
BITPIX = 32 / number of bits per data pixel
NAXIS = 2 / number of data axes
NAXIS1 = 2048 / length of data axis 1
NAXIS2 = 2048 / length of data axis 2
PCOUNT = 0 / required keyword; must = 0
GCOUNT = 1 / required keyword; must = 1
EXTNAME = 'Q1 '
HDUVERS = 1
DETSEC = '[2049:4096,1:2048]' / [px] section of DETSIZE
DATASEC = '[5:2044,5:2044]' / [px] section of CHIPSIZ
PERCT025= 2688. / 2.5 % percentile ADU
PERCT050= 2700. / 5 % percentile ADU
PERCT075= 2705. / 7.5 % percentile ADU
PERCT100= 2708. / 10 % percentile ADU
PERCT125= 2712. / 12.5 % percentile ADU
PERCT150= 2714. / 15 % percentile ADU
PERCT175= 2716. / 17.5 % percentile ADU
PERCT200= 2718. / 20 % percentile ADU
PERCT225= 2720. / 22.5 % percentile ADU
PERCT250= 2723. / 25 % percentile ADU
PERCT275= 2725. / 27.5 % percentile ADU
PERCT300= 2726. / 30 % percentile ADU
PERCT325= 2728. / 32.5 % percentile ADU
PERCT350= 2730. / 35 % percentile ADU
PERCT375= 2732. / 37.5 % percentile ADU
PERCT400= 2733. / 40 % percentile ADU
PERCT425= 2735. / 42.5 % percentile ADU
PERCT450= 2736. / 45 % percentile ADU
PERCT475= 2738. / 47.5 % percentile ADU
PERCT500= 2739. / 50 % percentile ADU
PERCT525= 2741. / 52.5 % percentile ADU
PERCT550= 2743. / 55 % percentile ADU
PERCT575= 2745. / 57.5 % percentile ADU
PERCT600= 2746. / 60 % percentile ADU
PERCT625= 2748. / 62.5 % percentile ADU
PERCT650= 2749. / 65 % percentile ADU
PERCT675= 2750. / 67.5 % percentile ADU
PERCT700= 2753. / 70 % percentile ADU
PERCT725= 2754. / 72.5 % percentile ADU
PERCT750= 2756. / 75 % percentile ADU
PERCT775= 2758. / 77.5 % percentile ADU
PERCT800= 2760. / 80 % percentile ADU
PERCT825= 2763. / 82.5 % percentile ADU
PERCT850= 2765. / 85 % percentile ADU
PERCT875= 2768. / 87.5 % percentile ADU
PERCT900= 2772. / 90 % percentile ADU
PERCT925= 2776. / 92.5 % percentile ADU
PERCT950= 2780. / 95 % percentile ADU
PERCT975= 2787. / 97.5 % percentile ADU
RA = 332.367528 / [deg] R.A.: 22:09:28.2
DEC = 51.084307 / [deg] Dec.: +51:05:04
PIXSCALE= 0.45 / [arcsec/px]
CUNIT1 = 'deg ' / WCS units along axis 1
CUNIT2 = 'deg ' / WCS units along axis 2
CTYPE1 = 'RA---TAN' / WCS axis 1
CTYPE2 = 'DEC--TAN' / WCS axis 2
CRVAL1 = 332.36752753434 / [deg] RA in mosaic center
CRVAL2 = 51.0843069975685 / [deg] DEC in mosaic center
CD1_1 = -0.000124999996688631 / [deg/px] WCS matrix diagonal
CD2_2 = 0.000124999996688631 / [deg/px] WCS matrix diagonal
CD1_2 = -2.56379278852432E-14 / [deg/px] WCS matrix outer diagonal
CD2_1 = -2.56379278852432E-14 / [deg/px] WCS matrix outer diagonal
CRPIX1 = -81 / [px] RA and DEC center along axis 1
CRPIX2 = 2132 / [px] RA and DEC center along axis 2
DET_ID = 'SG1 ' / lower right (SW) chip
COMMENT WCS assumes CHIPGAPX=167, CHIPGAPY=167, north=90 deg
BSCALE = 1.
BZERO = 0.
END
5.3. Observation Tool keywords¶
Next keywords are automatically added to the FITS header by the PANIC Observation Tool (OT), as each file is created. If these are not saved, neither PAPI nor PQL will work correctly:
OBS_TOOL= 'OT_V1.1 ' / PANIC Observing Tool Software version
PROG_ID = ' ' / PANIC Observing Program ID
OB_ID = '6 ' / PANIC Observing Block ID
OB_NAME = 'OB CU Cnc Ks 2' / PANIC Observing Block Name
OB_PAT = '5-point ' / PANIC Observing Block Pattern Type
PAT_NAME= 'OS Ks 2 ' / PANIC Observing Secuence Pattern Name
PAT_EXPN= 1 / PANIC Pattern exposition number
PAT_NEXP= 5 / PANIC Pattern total number of expositions
IMAGETYP= 'SCIENCE ' / PANIC Image type
5.4. Data¶
Raw images pixels are coded with 32-bit signed integers (BITPIX=32), however final reduced images are coded with 32-bit single precision floating point (BITPIX=-32). The layout of each chip image in a raw image is described above.
5.5. Classification¶
Any raw frame can be classified on the basis of a set of keywords read from its header. Data classification is typically carried out by the Pipeline at start or by PQL, that apply the same set of classification rules. The association of a raw frame with calibration data (e.g., of a science frame with a master dark frame) can be obtained by matching the values of a different set of header keywords (filter, texp, ncoadds, itime, readmode, date-obs, etc). Each kind of raw frame is typically associated to a single PAPI pipeline recipe, i.e., the recipe assigned to the reduction of that specific frame type. In the pipeline environment this recipe would be launched automatically. In the following, all PANIC raw data frames are listed, together with the keywords used for their classification and correct association.
Type |
Description |
|---|---|
|
Dark frame |
|
Dome flat-field frame (lamp on/lamp off) |
|
Sky flat-field frame |
|
Focus frame of a focus series |
|
Science frame |
|
Sky frame (mostly clear) used for extended object reduction |
5.6. Data grouping¶
Once the raw files are classified, they are grouped into observing sequences, taking into account the keywords added by the Observation Tool (OT), and finding out the dither sequences observed. This way, all files beloging to the same observing sequence will be processed together.